According to the World Health Organization (WHO), 3.6 billion medical imaging tests are performed every year globally to diagnose, monitor, and treat various conditions. Most of these images are stored in a globally recognized standard called DICOM (Digital Imaging and Communications in Medicine). Imaging studies in DICOM format are a combination of unstructured images and structured metadata.
Typical data management systems such as data warehouses do not accommodate unstructured data types. And data lakes fail to catalog and store metadata, which is critical for search, governance and accessibility of these imaging exams. Databricks Pixels 0.6, developed in 2021, addressed many of these challenges by providing a scalable environment from which you can ingest, manage, and catalog all of your medical imaging data within the Databricks Data Intelligence Platform.
Now, with Databricks Pixels 2.0 Solution Accelerator, additional enhancements include integrations with NVIDIA accelerated computing platforms and MONAI. MONAI is a set of open-source frameworks for accelerating research and clinical collaboration in medical imaging. This integration offers considerable improvements, including end-to-end capabilities for ingesting, managing, and analyzing healthcare images that can meaningfully assist clinical analysis.
This post walks through the benefits of these integrations and how to quickly develop a proof of concept application using Pixels 2.0 that displays CT studies, pre-annotates them using AI, enables users to make corrections, and then fine-tunes the model with any updates in real time (active learning).
AI-powered medical image processing
One of the most significant advancements in healthcare has been the integration of AI into medical imaging. AI-powered systems are transforming radiology by streamlining workflows, reducing workloads for radiologists, and improving patient outcomes. These technologies can detect abnormalities in imaging studies, prioritize urgent cases, and enable faster diagnoses and treatment planning. This is particularly critical in addressing the growing demand for imaging services and the shortage of radiology professionals.
However, delivering on these promises requires the ability to consolidate and manage diverse data sources, including imaging files, electronic health records (EHRs), radiology reports and clinical data. Furthermore, the integration of AI in medical imaging poses additional challenges, such as managing complex MLOps workflows for model training and operationalization, while ensuring compliance with stringent regulations like HIPAA and GDPR. Effective governance, visibility, and access to data are essential to overcome these challenges.
Challenges of using DICOM for life sciences analytics
DICOM is a global healthcare standard that describes the structure and transport of medical images between different systems, such as X-ray equipment, storage systems, and medical-grade viewers.
Broadly speaking, a DICOM file contains a header with rich metadata information and a set of one or more frames of image intensity values (pixels). The tags, while arranged in a complex structure, contain valuable information, and are indexed in their entirety. Other solutions often pull a small subset of the tags.
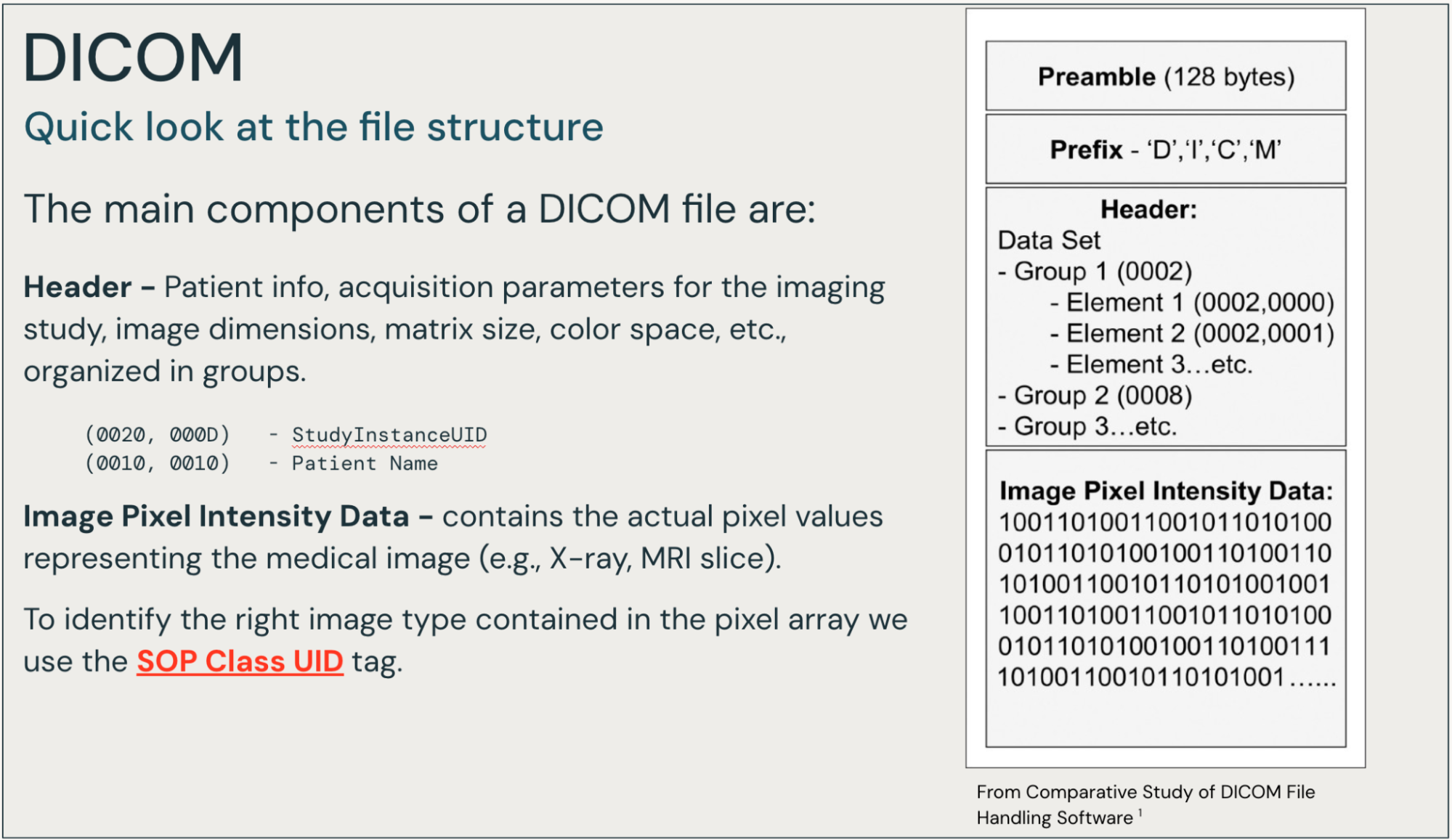
In our exploration of healthcare and life sciences analytics workflows involving DICOM and other medical images, we’ve observed a common challenge. Many organizations face a landscape of disconnected technology solutions, often lacking cohesive governance in terms of access controls, audit logs, and data lineage.
It’s not uncommon to find a single organization where multiple research groups have independently developed their own approaches for ingesting and preparing DICOM files for analysis and modeling. This fragmentation typically results in:
- 5-10 distinct groups of researchers
- 5-10 different solutions for handling DICOM files
- Multiple technologies in use across the organization
- Significant IT resources devoted to isolated data management tasks
To fully realize the potential of AI in medical imaging, tools that simplify the management of imaging data, like DICOM, are critical.
Impact on research teams
This technological fragmentation presents significant challenges for various team members, including bioinformaticians, data engineers, and data scientists. These professionals often find themselves grappling with the complexities of scaling end-to-end processing workflows. The lack of a unified, streamlined approach can impede efficiency and potentially slow down valuable research progress. By addressing these challenges and working towards more integrated, governed solutions, organizations have the opportunity to significantly enhance their research capabilities and outcomes in the field of medical imaging analytics.
The integration of Databricks Pixels 2.0 Solution Accelerator with NVIDIA accelerated computing platforms and MONAI aims to empower individuals across the healthcare industry, from researchers to analytics professionals to data scientists. The benefits include the following:
- Accelerated research: Researchers can develop and train AI models for medical imaging faster than ever before.
- Improved diagnostic accuracy: AI-assisted image analysis can help radiologists identify abnormalities with greater precision.
- Streamlined workflows: The Solution Accelerator automates time-consuming tasks, allowing healthcare providers to focus more on patient care.
- Enhanced collaboration: The platform facilitates easier sharing of insights and models among healthcare institutions, fostering innovation in the field.
The ability to unify and govern all modalities of healthcare datasets on the Databricks Data Intelligence Platform—including HL7 and FHIR, DICOM, and more—helps to optimize analytically driven workflows.
Databricks ingests, processes, and stores derived metadata, features, and segments on to your cloud storage account. This Solution Accelerator performs indexing, de-identification, and featurization with machine learning (ML) models like the MONAI Label Auto Segmentation model. It also performs interactive ML active learning workflows leveraging labeling and rich visualizations. All of these activities are secured in a HIPAA-compliant and scalable cloud environment required to reliably process anywhere from hundreds to billions of DICOM images.
The evolution of the Databricks Pixels Solution Accelerator
The core intent of the first edition of Databricks Pixels 0.6 Solution Accelerator was to accelerate time to value for the ingestion, indexing, and accessibility of DICOM metadata as part of the Healthcare and Life Science Lakehouse. According to Douglas Moore, the initial author of the Pixels Solution Accelerator, “Running SQL over DICOM metadata for customers was a compelling vision.”
Pixels 0.6 uses off-the-shelf and well-tested Pydicom and GDCM libraries to open the DICOM file to extract the header metadata tags, all of the tags, and a few metrics from the image. These operations were scaled up and out with Spark User Defined Functions (UDFs), while the cloud layer was abstracted away with a FUSE-based DBFS mount or S3FS API calls.
Databricks customers de-silo data. A Lakehouse architecture enables easy integration of DICOM derived from EHRs, claims, and genomic data sets. UC Davis Health, for example, has seen tremendous benefit from the use of Pixels.
According to Peter Paing Soe, enterprise data architect at UC Davis Health, “We use the Pixels Solution Accelerator to ingest DICOM images into our Databricks environment. Our unified Lakehouse platform provides faculty and staff with integrated access to comprehensive clinical data and DICOM images, paired with effective Databricks computational resources.”
The collaboration between Databricks and NVIDIA
The Databricks Data Intelligence Platform offers scalable solutions for data together with AI processing to harness the power of AI in medical imaging. Databricks provides extensive governance, data processing, and a broad base of AI services.
NVIDIA delivers accelerated computing (GPUs) alongside high-quality pretrained models (such as MONAI) tailored for medical imaging workflows. NVIDIA is the primary sponsor of the MONAI and Open Health Imaging Foundation (OHIF) communities.
Pixels 2.0 Medical Imaging Solution Accelerator brings together Databricks and NVIDIA components into a package that provides a reference implementation and a well-governed reference architecture. Pixels 2.0 is installed in minutes, running within the hour.
The net result of this collaboration between Databricks and NVIDIA is accelerated time to value for optimizing medical imaging workflows.
Key features of Databricks Pixels 2.0 Solution Accelerator
Pixels 2.0 provides the following key capabilities:
- Streaming, incremental batch, and full historical load and processing: Ingest, unzip, index, and perform de-identification, AI-based segmentation, and additional featurization on a full historical load basis, on an incremental batch basis (for example, every day or hour) or as a continuous stream.
- Unified governance, data sharing with Unity Catalog: Govern the raw data, the complex structured data from the tags, derived aggregates and cohorts, and the AI models.
- Protected Health Information (PHI) redaction: De-identify PHI tags and image data through open-source or commercially available packages.
- Scale to zero model serving, inference, segmentation, and active learning: Cost effectively applying AI and ML in a production environment, processing an archive of DICOMs, daily batch processing, hourly mini-batches, streaming, or interactive needs driven by user-oriented applications.
- Interactive OHIF viewer with labeling integrated as an integrated Lakehouse App: In human-centered workflows, the ability to visualize, label, and command ML operations on images stored in the Lakehouse.
- Open APIs and Delta Sharing, Clean Rooms: Power interoperability among departments and devices. Fosters open (and secure) collaboration among different organizations.
The reference solution architecture diagram in Figure 2 summarizes the capabilities brought together by Databricks Pixels 2.0 Solution Accelerator.
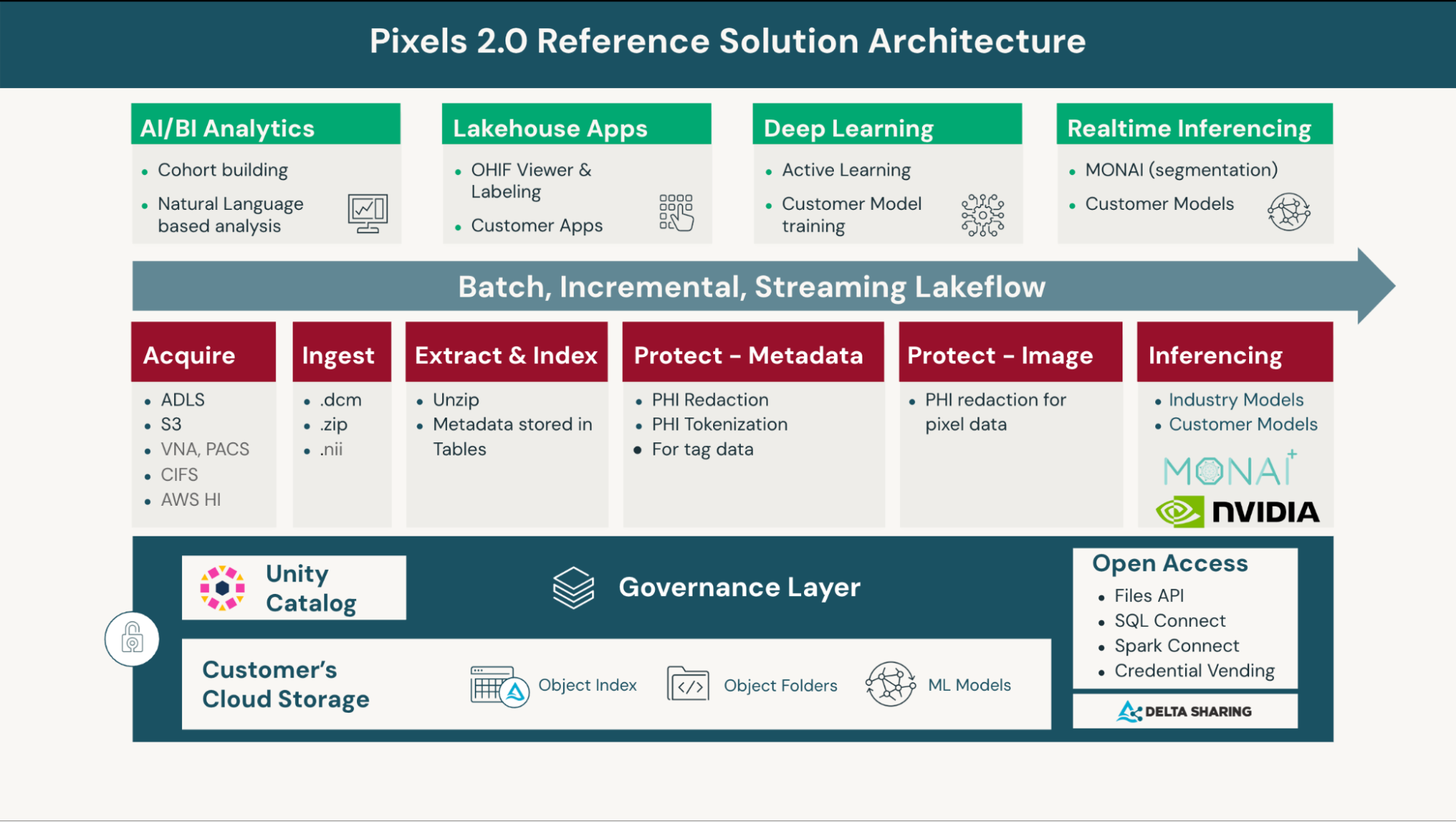
By bringing together all of these capabilities into one Solution Accelerator, organizations can achieve the needed workflow optimizations, reduce complex architectures, and achieve the scale they need.
Efficiently process and analyze medical imaging data
Used together, Databricks and MONAI are able to address one of the most pressing challenges in healthcare: efficiently processing and analyzing the vast amounts of medical imaging data generated daily.
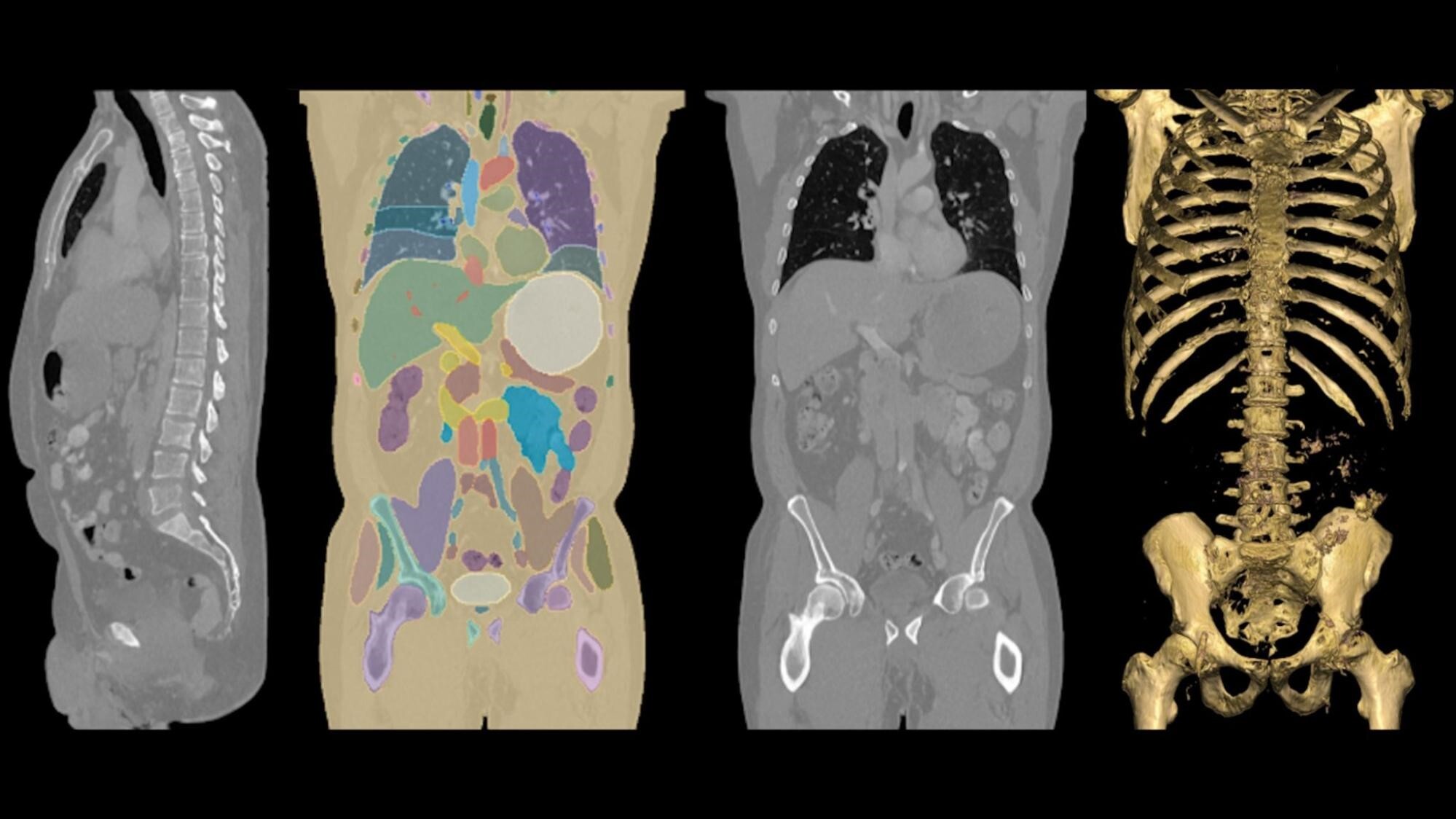
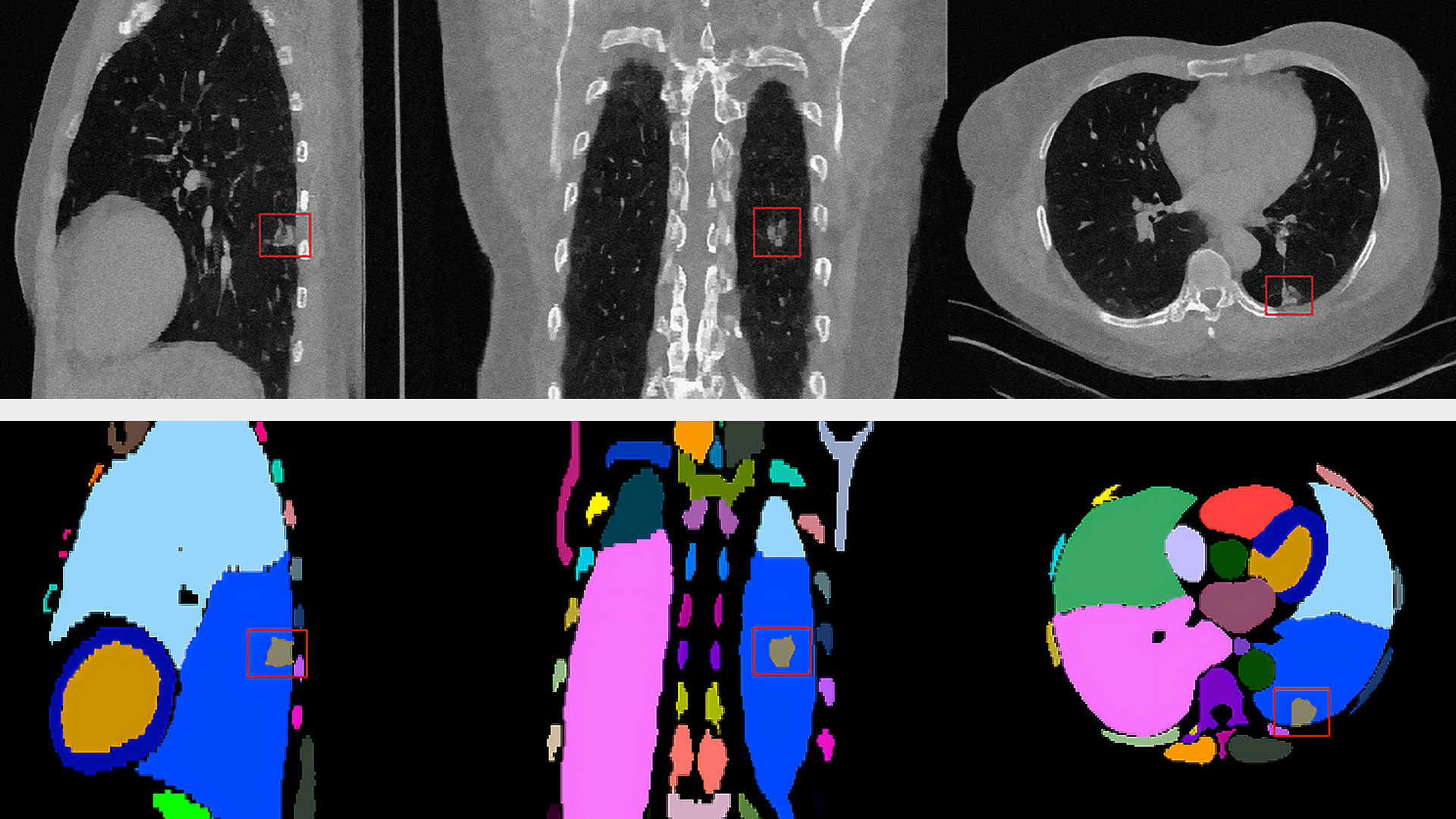
MONAI Label is an intelligent tool for creating, training, and deploying ML models for medical image annotation and segmentation. It reduces the time and effort required for data labelling by up to 75% using active learning. The tool facilitates automatic segmentation of pixels and voxels within CT scans. Inference over an imaging study with more than 1,000 DICOM image frames results in a detailed, color-coded overlay and precise vector representation of the organs within CT scans of the human torso.
This effort introduces production-scale auto segmentation of CT images in batch, streaming, and real-time inference modes. The model (out-of-the-box or fine-tuned) is registered into Databricks Unity Catalog. The job at run time loads the model and weights, then performs inferencing on DICOM files.
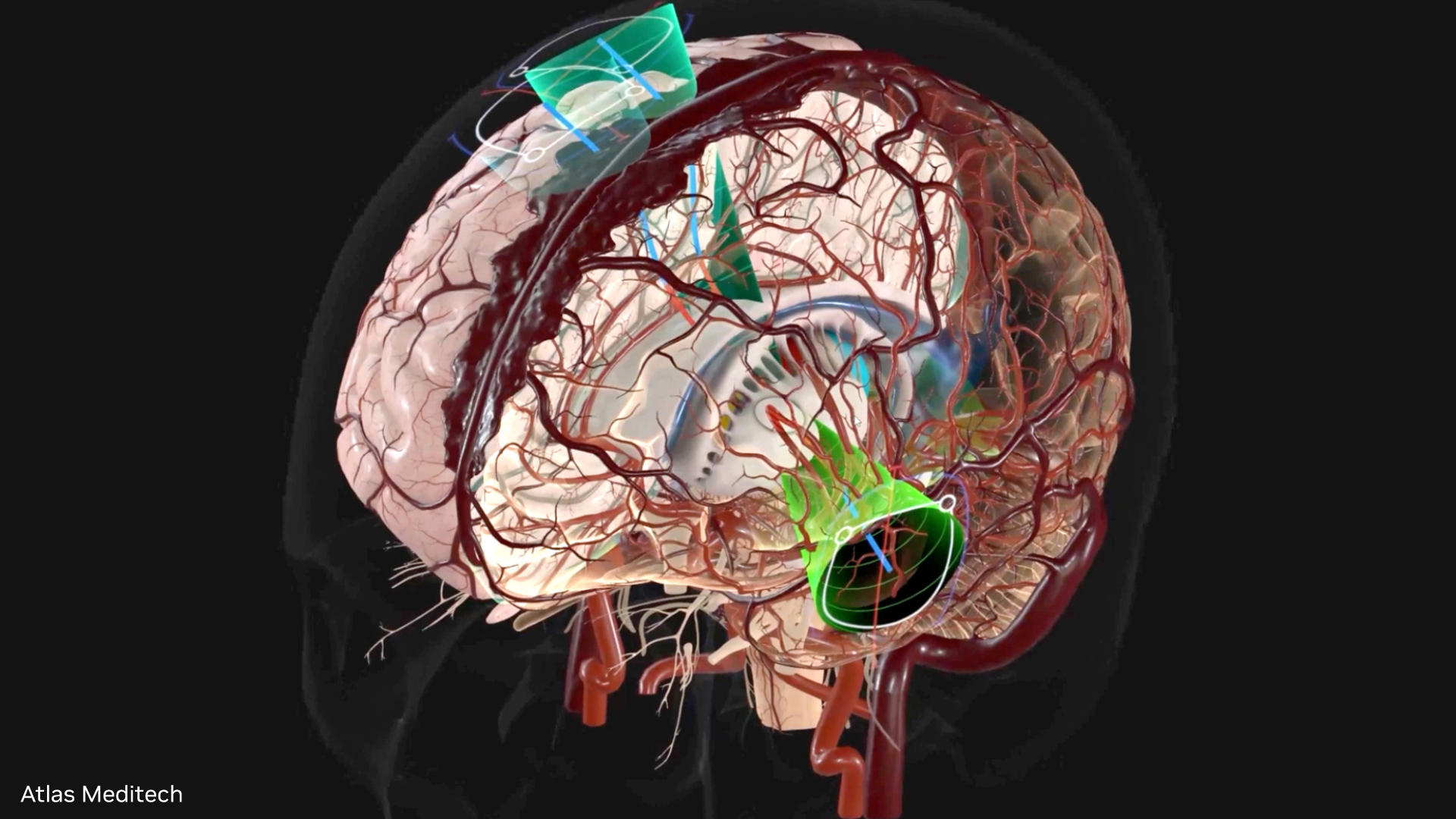
For interactive use cases, MONAI Label is deployed to a GPU-based, scale-to-zero endpoint. The model endpoint is fully managed, and new “champion” versions of the model are automatically deployed to production. The secured model serving endpoint makes building interactive data apps easy. For example, the OHIF viewer, a medical-grade open source imaging viewer, is easy to integrate and govern.
The active learning workflow involves labeling portions of the CT Scan, saving the annotations (labels) back into Databricks, and then retraining the model on a Databricks-managed machine learning GPU cluster. The active learning workflow is entirely driven by interacting with the OHIF viewer. The OHIF viewer is integrated into the Databricks Data Intelligence Platform security “umbrella” as a Lakehouse App.
Get started
The future of healthcare is data-driven. The integration of Databricks Pixels 2.0 solution with NVIDIA accelerated computing platforms and MONAI offers considerable improvements, including end-to-end capabilities for ingesting, managing, and analyzing healthcare images that can meaningfully assist clinical analysis.
You can quickly develop a proof of concept application using Pixels 2.0 that displays CT studies, pre-annotates them using AI, enables users to make corrections, and then fine-tunes the model with any updates in real time (active learning).
Ready to get started exploring the Databricks-NVIDIA Solution Accelerator? Follow the steps below:
- Log in to your Databricks workspace or create a new trial with the express setup experience. Select ‘Professional’.
- Fork the GitHub repo, and clone into a repo folder in your Databricks workspace.
- Run the RUNME notebook on a Databricks cluster to create an example ingestion and segmentation pipeline.
- Go through the additional notebooks called out in the README to deploy your Lakehouse App or active learning setup.
For additional support, reach out to your Databricks or NVIDIA account team. You can also visit databricks-industry-solutions/pixels on GitHub to post questions and issues. To learn more about MONAI, check out the MONAI Quickstart Guide and MONAI Model Zoo.